PATHWAY-BASED SAFETY ASSESSMENT FAQs
In brief, this approach involves a characterization of the inherent properties of a chemical (or drug – a drug is a chemical with special properties!), and the application of our knowledge of the underlying biology – biological pathways (that’s where the “pathway-based” comes from), combined with an understanding or estimate of exposure, to design a testing strategy that will give exactly the information needed to safely use that chemical (or drug). Sound fantastical? That’s what pretty much everybody thought when the National Academy of Sciences proposed it in 2007 (NRC, 2007), but it turns out that it’s not so far-fetched after all, and enormous progress has been and continues to be made since then.
Chemical characterization
First, our ability to understand how chemical structure is related to physical or biological properties has greatly improved (from simple things like water solubility and acidity to more complicated things like whether it might be an allergen), as has our ability to recognize similarities between chemicals, and when that similarity might indicate similar properties.
Our ability to test biological properties of chemicals using cell extracts, or cell- or tissue-based testing has also greatly improved, and is the focus of several large-scale projects, including the US National Center for Translational Sciences (NCATS)/National Institutes of Health (NIH)/Environmental Protection Agency (EPA)’s Tox21 program, which applies hundreds of cell based tests to look at thousands of biological activities of thousands of chemicals. The data generated from this program is publicly available. The EPA and NIH continue to develop computer-based models and interpretation methods to use this information in chemical safety decisions.
Biological knowledge and systems biology
One of the great shortcomings in our ability to ameliorate disease or protect from adverse chemical exposure is our lack of understanding of the underlying biology involved. And yet, we’ve seen an explosion in biological research over the past several decades – we know more about biology – of animals, of humans, of everything, than ever before – so what’s the deal? One of the issues is how we store our biological information. Most everything is in papers, or reports, or databases that are all separate and use different formats or software, so that this information doesn’t actually coalesce into common “knowledge” about biology. The “silo-ing” of information is further compounded by specialization of science – scientists generally spend their careers focused in one area of one field – and since there is so much information in that relatively small area of biology, it’s not feasible to venture out to gain a wider perspective. So how do we surmount these siloes and bring together all of the information that might be held in these different places, formats, or individual knowledge, into a common platform, using a common language, so that it becomes collective knowledge that can be used to improve our understanding of disease and the potential for chemical interference? Here’s were systems biology – the explicit understanding of network of biological pathways – comes into the picture. Data-driven discovery of biological pathways infer pathways from molecular associations, whereas curated pathway frameworks are assembled and assessed manually from data sources, including published literature. These varied approaches to systems biology serve different functions, the first to discover previously unknown pathways and associations, the second to form a reliable basis upon which to found hypothesis-driven research or regulatory decisions (Marshall et al., 2018). As an example, improved understanding of the complexities of cancer through increased levels of data integration is the goal of the Cancer Systems Biology Consortium, a community of experimental biologists and computer modelers integrating data from a variety of sources, including clinical samples, single cell analyses, network inference, image analysis, machine learning, and evolutionary theory, to build, test, and validate hypotheses and ideas (Marshall et al., 2018).
Application of systems biology to toxicology: mode-of-action and adverse outcome pathways and systems toxicology
Recent global activities have been focused on initiating a paradigm shift in the way in which drug and chemical toxicity characterizations are conducted; with one major area being the definition and use of pathway-based assessments (Willett et al., 2014). Pathway-based assessments involve the prediction of outcomes in the organism or population based on measurement of upstream biological mechanisms at the molecular, cellular and tissue levels (See, AOPs in the Get Trained section). To this end, several overlapping and related frameworks have been proposed, including modes of action (MoA), adverse outcome pathways, toxicity pathways and pathways of toxicity.
The following paragraphs provide a better overview of the history of pathway development, MoA frameworks, AOPs, and systems toxicology.
The idea of incorporating mechanistic biochemical information into toxicological assessment is not new; it began with dose-response modeling efforts (e.g. Clewell et al., 1995) and mode-of-action frameworks, such as those developed by the International Program on Chemical Safety (IPCS) to determine the human relevance of modes-of-action of pesticides and industrial chemicals leading to carcinogenicity (and later non-carcinogenic toxicity) (Boobis et al., 2006; Boobis et al., 2008), and the creation of mode of action pathways commonly used in drug development and applied to human disease. The notion of toxicity pathways as articulated by the National Research Council in 2007 in its report (NRC, 2007), Toxicity Testing for the 21st Century: a Vision and a Strategy, takes this concept a bit further by envisioning a system-wide network of pathways that leads to a predictive, hypothesis-driven assessment paradigm for toxicity in general. The goals of this new approach are to improve efficiency and decrease uncertainty in risk and hazard evaluations. In 2012, this concept has been further formalized for toxicological assessment for both human health and ecological endpoints as the Adverse Outcome Pathway (AOP) and has been taken up by the Test Guidelines Program at the Organization for Economic Cooperation and Development (OECD) as an organizing principle for all test guidelines.
The IPCS cancer and non-cancer MoA frameworks outline a systematic process of describing chemical MoA in animals and comparing those with likely MoA in humans to determine human relevance (Meek et al., 2003). Several founding principles of pathway-based approaches are articulated in the original IPCS frameworks: MoA defined as a series of key events along a biological pathway from the initial chemical interaction through to the toxicological outcome; a recognition that a MoA does not need to be complete to be useful and that its use depends on level of completeness (e.g., incomplete MoA can inform testing strategies but is likely not sufficient to support hazard classification); a focus on a single MoA at a time while recognizing that a chemical may have more than one MoA and that several modes may be related; definition of a “key event” as a step in the pathway that is critical to development of the toxicological outcome and is measurable; a requirement to systematically establish causation between key events; the importance of establishing quantitative parameters in order to apply the MoA to risk assessment; and the need to establish relevance to human biology. In this context, mode-of-action is distinguished from mechanism of action: the latter being defined as a more detailed description of the pathway that includes molecular interactions.
Establishing evidence for the MoA hypothesis is based on the Bradford-Hill criteria for establishing causation: strength of association, consistency of the evidence, specificity of the relationship, consistent temporal relationships, dose-response relationships, biological plausibility, coherence of the evidence, and consideration of alternative explanations (Bradford-Hill, 1965). The IPCS frameworks recommend determining human relevance by answering four key questions: 1) is there sufficient weight-of-evidence for the MoA in animals? 2) can human relevance be excluded on the basis of qualitative differences in key events? 3) can human relevance be excluded on the basis of quantitative differences in key events? and 4) do the quantitative differences affect the default uncertainty factors applied in risk assessment?
The MoA framework has been updated to accommodate insights from the expanding application of pathway-based approaches to risk assessment in general (Meek et al., 2014). In this framework, MoA and AOP are conceptually similar, with a distinction in that MoA does not necessarily imply adversity; it can also refer, for example, to efficacy of a drug.
The term systems toxicology was coined to describe the application of systems biology approaches to toxicological studies (Mc Auley et al., 2015). In practice, systems toxicology is the integration of classical toxicology with quantitative analysis of large networks of molecular and functional changes occurring across multiple levels of biological organization (Sturla et al., 2014). This multidisciplinary approach is expected to create knowledge regarding both the dynamic interactions between biomolecular components of a complex biological system and how perturbing these interactions with substances alters homeostasis and leads to adverse reactions and disease (Sturla et al., 2014).
The AOP framework is a way to formalize systems toxicology and provides a platform for explicitly describing and evaluating the state of knowledge of the pathways that make up biological networks. An AOP describes the events that occur following chemical exposure, beginning with the molecular interaction of the chemical with a biomolecule (e.g., a protein, receptor, etc.) – the molecular initiating event (MIE) – followed by a description of the sequential cellular and tissue perturbations (Key Events, KEs) that lead to eventual toxicological effect – or adverse outcome (Figure 1). Although not traditionally associated with systems toxicology, as more and more AOPs are developed, the increasing complexity of the AOP network will approach systems toxicology. In addition, as more detailed causal and quantitative information is included in the relationships between KE, AOPs will increasingly support predictive modeling. When AOPs containing quantitative information are combined with exposure metrics and toxicokinetics from PBPK models, complete predictive models can be built. Overall, the development of dynamic (executable) mathematical models of AOPs linking exposure to individual and population-wide adverse outcomes is the grand challenge of computational systems toxicology, where the systems description should enable prediction of responses for which experimental data were not available (i.e., the system will exhibit emergent properties entailing novel patterns and properties arising from the inherent structure of the system) (Sturla et al., 2014).
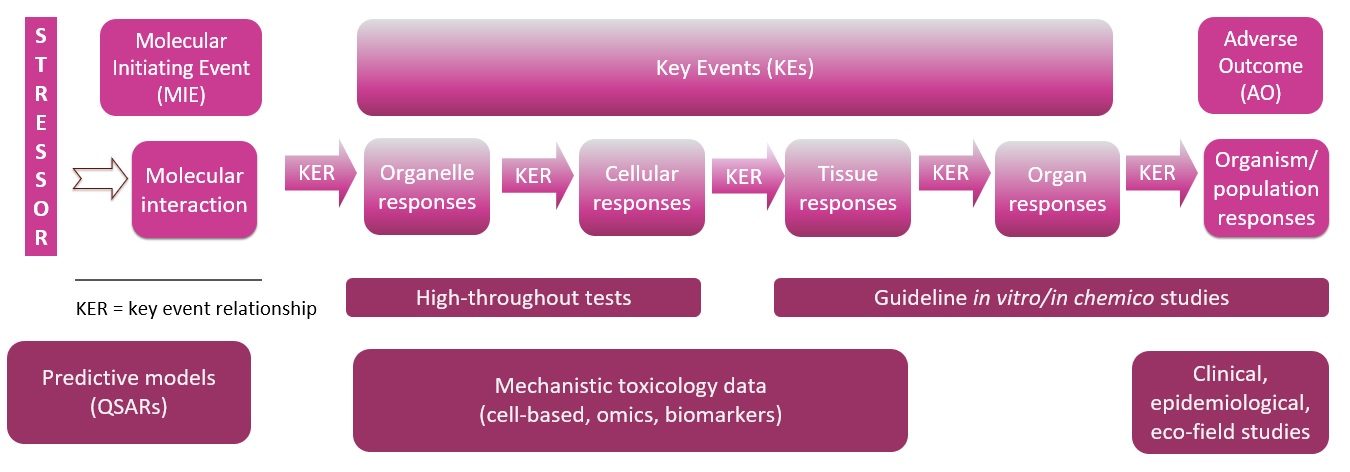
Figure 1. Diagram of an AOP
Advances in biological understanding as well as in experimental technologies (New Approach Methodologies or NAMs, e.g. ‘omics tools, stem cell culturing, reconstructed tissues), have allowed the contemplation of a dramatically different approach to understanding disease and toxicology than those traditionally practiced. This approach couples bespoke testing strategies using NAMs with existing knowledge of normal biology and the consequences of disturbing that biology. This allows a structured, transparent, and hypothesis-based approach to predicting adverse outcomes from the measurements of upstream perturbations at the cellular or tissue level.
Taking into consideration our current understanding of biology, we can use a large collection of NAMs (high- or medium-throughput) to rapidly characterize or “finger print” one or several (hundreds or even thousands) chemicals. This gives us a picture of the overall level of biological activity of each chemical as well as an initial idea of what type of biological activity a chemical might have (which biological pathways it potentially perturbs). The most biologically active chemicals are those that are most likely to pose a concern when living systems are exposed to them. We can then select the most active chemicals for further study and, again using our understanding of the biology of the underlying pathways, begin to design testing strategies to further characterize the type of potential activity.
Innovative research in cell and tissue culturing is also progressing and is aiming at developing “organ-on-a-chip” microsystems and models that will provide further insights of chemical’s toxicity and exposure in the human body, also feeding into innovative computational systems toxicology models. The ability to use human-derived stem cells is also advancing for the creation of novel, in vitro models to explore several biological processes, but also in clinical applications. Overall, the future of toxicology and health research is expected to proceed toward a human-focused paradigm, in line with the National Research Council’s vision for Toxicity Testing in the 21st Century.
The new way of thinking about chemical safety decisions involves the use of hypothesis-based integrated testing strategies, or IATA(Integrated Approaches to Testing and Assessment). OECD guidance on developing IATA defines IATA as “pragmatic, science-based approaches for chemical hazard or risk characterization that rely on an integrated analysis of existing information in a weight of evidence assessment coupled with the generation of new information using testing strategies…IATA follow an iterative approach to answer a defined question in a specific regulatory context…” (Sachana and Leinala, 2017; OECD, 2016). The IATA framework is flexible and allows streamlining of the testing and assessment process to generate exactly the information needed to make a decision. IATA can be as simple or as complex as needed to answer the specific question, depending on the amount of certainty needed. IATA begin with the problem formulation – what is the question that needs to be answered and what is the context of that question. Here is where potential use and exposure to the chemical is considered. The next step is to gather all existing information, apply a weight of evidence consideration to that information, and ask whether the question has been answered with sufficient certainty. If not, a testing strategy may be considered. Here is where AOPs, or systems toxicology, can contribute. Biological knowledge is used to design a hypothesis-based test strategy, using NAMs designed to provide the appropriate information. The results from these tests are again evaluated by weight-of-evidence to see if the question has now been answered with enough confidence; this process is repeated until it has.
In this way, by designing testing strategies around the emerging pathway- and network knowledge, it is possible to converge with the systems understanding and providing the data for its modeling (Hartung et al., 2017). Systems toxicology is expected to enable the shift from toxicological assessment using solely apical endpoints toward a pathway-based approach to safety testing.
The AOP concept is useful in developing a predictive toxicological framework in several ways. Even incomplete AOPs can inform chemical grouping or categories and structure activity relationships, they can aid in increasing certainty of interpretation of both existing and new information, they can identify alternative endpoints for use in hazard assessment, and, as mentioned before, they can be used to inform IATAs and integrated testing strategies (ITS) that maximize useful information gained from minimal testing (Willett et al, 2014). Furthermore, the increasingly growing knowledge about biological and disease pathways is expected to allow for development of more complex and comprehensive networks from individual AOPs, thus better informing future research and predictive and regulatory toxicology. The resulting AOP networks can subsequently be analyzed in a variety of ways to extract useful information, including topological analyses, critical pathway identification, and characterization of interactions among AOPs within a network (Knapen et al., 2018). As a natural evolution of qualitative AOPs, it is expected that more quantitative AOPs (qAOPs) will be developed, thus providing quantitative, dose-response and time-course predictions that can be considered for risk assessment purposes. There is also a growing consensus that AOPs will best serve toxicology if methods can be found to integrate dose–response relationships and toxicokinetics with AOPs to build the concept of ‘source to outcome pathways’ (Sewell et al., 2018), therefore integrating the aggregate exposure pathway (AEP) and the AOP frameworks. This should result into a more holistic perspective, that can further support regulatory-decision making.